# load packages
library(countdown)
library(tidyverse)
library(lubridate)
library(janitor)
library(colorspace)
library(broom)
library(fs)
# set theme for ggplot2
ggplot2::theme_set(ggplot2::theme_minimal(base_size = 14))
# set width of code output
options(width = 65)
# set figure parameters for knitr
knitr::opts_chunk$set(
fig.width = 7, # 7" width
fig.asp = 0.618, # the golden ratio
fig.retina = 3, # dpi multiplier for displaying HTML output on retina
fig.align = "center", # center align figures
dpi = 300 # higher dpi, sharper image
)Visualizing time series data II
Lecture 10
University of Arizona
INFO 526 - Fall 2023
Warm up
Announcements
- Don’t forget to watch the videos for Wednesday’s lecture
Project next steps I
- Proposal feedback is available.
Required: Address all issues and close them.
Optional: Submit for regrade, by replying to the “Proposal feedback” issue and tagging me in your reply. Due 5pm tomorrow (Tuesday). Final proposal score will be midpoint of original and updated score.
Project next steps II
- Create a slide deck in
presentation.qmd, replacing the placeholder text. - Add your write-up to
index.qmd, replacing the placeholder text. - In both outputs, hide the code with
echo: falseprior to turning them in. - Due: Wednesday, Monday, October 16th, 3pm.
Setup
Working with dates
AQI levels
The previous graphic in tibble form, to be used later…
AQI data
Source: EPA’s Daily Air Quality Tracker
2016 - 2022 AQI (Ozone and PM2.5 combined) for Tucson, AZ core-based statistical area (CBSA), one file per year
2016 - 2022 AQI (Ozone and PM2.5 combined) for San Francisco-Oakland-Hayward, CA CBSA, one file per year
2022 Tucson, AZ
# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source
<date> <dbl> <chr> <chr> <chr> <chr>
1 2022-01-01 40 PM2.5 GERONIMO 04-019-11… AQS
2 2022-01-02 55 PM2.5 GERONIMO 04-019-11… AQS
3 2022-01-03 55 PM2.5 GERONIMO 04-019-11… AQS
# ℹ 362 more rows
# ℹ 5 more variables: x20_year_high_2000_2019 <dbl>,
# x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>Visualizing Tucson AQI
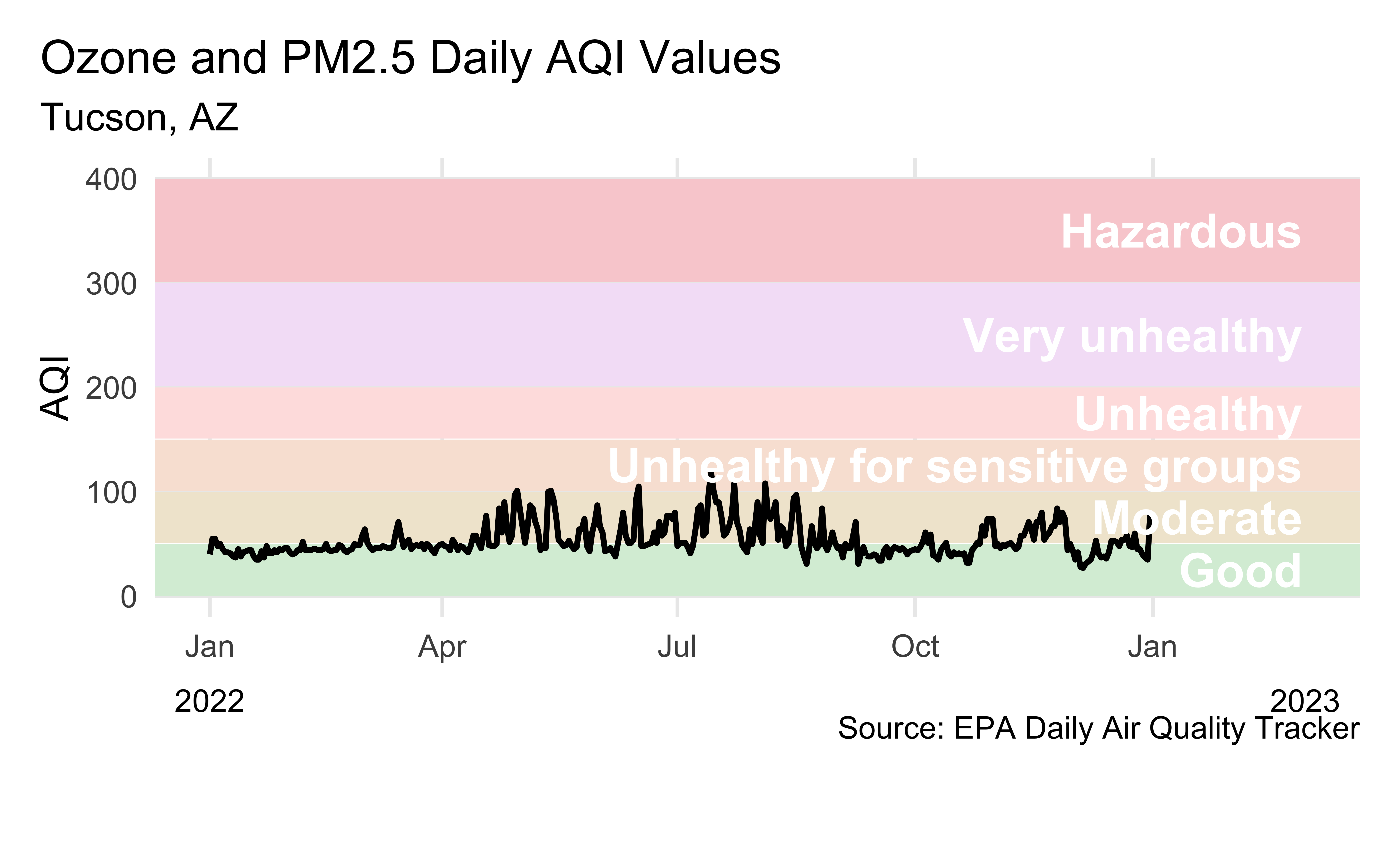
Another visualization of Tucson AQI
Recreate the following visualization.
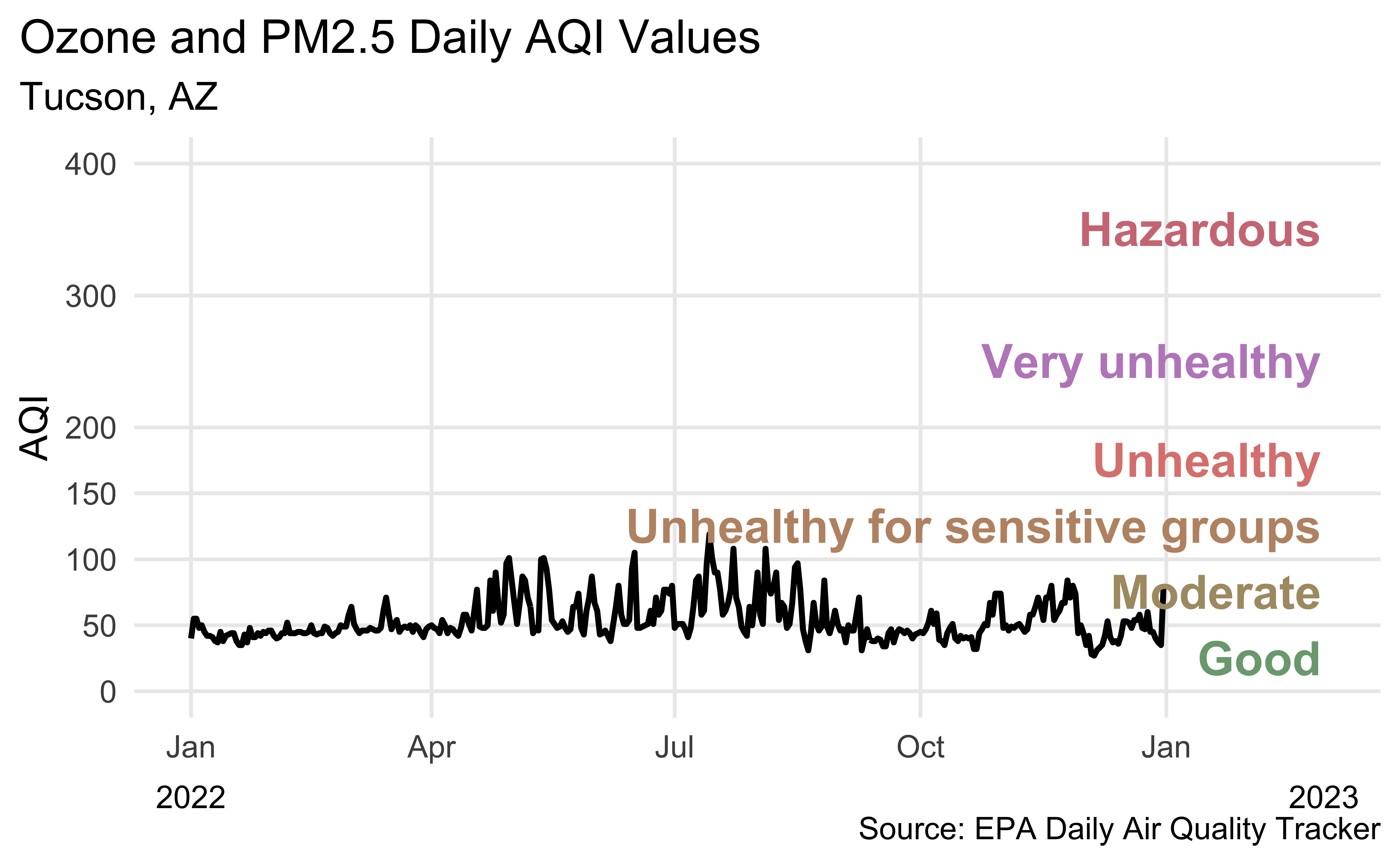
Highlights
The lubridate package is useful for converting to dates from character strings in a given format, e.g.
mdy(),ymd(), etc.The colorspace package is useful for programmatically darkening / lightening colors
scale_x_date: Setdate_labelsas"%b %y"for month-2 digit year,"%D"for date format such as%m/%d/%y, etc. See help forstrptime()for more.scale_color_identity()orscale_fill_identity()can be useful when your data already represents aesthetic values that ggplot2 can handle directly. By default doesn’t produce a legend.
Calculating cumulatives
Cumulatives over time
When visualizing time series data, a somewhat common task is to calculate cumulatives over time and plot them
In our example we’ll calculate the number of days with “good” AQI (≤ 50) and plot that value on the y-axis and the date on the x-axis
Calculating cumulatives
Step 1. Arrange your data
Calculating cumulatives
Step 2. Identify good days
tuc_2022 |>
select(date, aqi_value) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(good_aqi = if_else(aqi_value <= 50, 1, 0))# A tibble: 365 × 3
date aqi_value good_aqi
<date> <dbl> <dbl>
1 2022-01-01 40 1
2 2022-01-02 55 0
3 2022-01-03 55 0
4 2022-01-04 48 1
5 2022-01-05 50 1
# ℹ 360 more rowsCalculating cumulatives
Step 3. Sum over time
tuc_2022 |>
select(date, aqi_value) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(
good_aqi = if_else(aqi_value <= 50, 1, 0),
cumsum_good_aqi = cumsum(good_aqi)
)# A tibble: 365 × 4
date aqi_value good_aqi cumsum_good_aqi
<date> <dbl> <dbl> <dbl>
1 2022-01-01 40 1 1
2 2022-01-02 55 0 1
3 2022-01-03 55 0 1
4 2022-01-04 48 1 2
5 2022-01-05 50 1 3
# ℹ 360 more rowsPlotting cumulatives
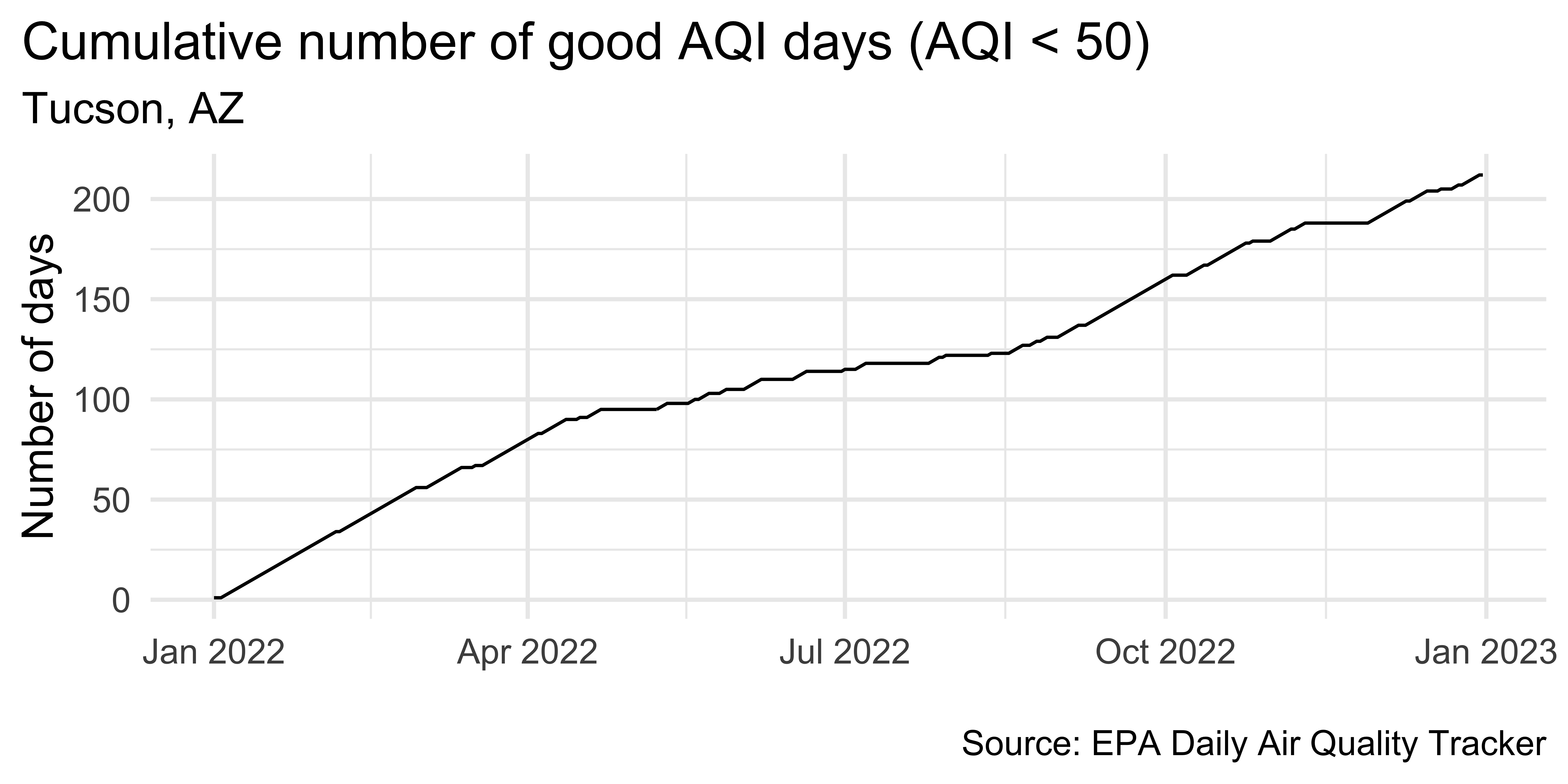
tuc_2022 |>
select(date, aqi_value) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(
good_aqi = if_else(aqi_value <= 50, 1, 0),
cumsum_good_aqi = cumsum(good_aqi)
) |>
ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) +
geom_line() +
scale_x_date(date_labels = "%b %Y") +
labs(
x = NULL, y = "Number of days",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "Tucson, AZ",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")Detrending
Detrending
Detrending is removing prominent long-term trend in time series to specifically highlight any notable deviations
Let’s demonstrate using multiple years of AQI data
Multiple years of Tucson, AZ data
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2017.csv
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2018.csv
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2019.csv
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2020.csv
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2021.csv
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2022.csv
/Users/gchism/Desktop/INFO526-F23/slides/09/data/tucson/ad_aqi_tracker_data-2023.csvReading multiple files
tuc <- read_csv(tuc_files, na = c(".", ""))
tuc <- tuc |>
janitor::clean_names() |>
mutate(
date = mdy(date),
good_aqi = if_else(aqi_value <= 50, 1, 0)
) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(cumsum_good_aqi = cumsum(good_aqi), .after = aqi_value)
tuc# A tibble: 2,392 × 13
date aqi_value cumsum_good_aqi main_pollutant site_name
<date> <dbl> <dbl> <chr> <chr>
1 2017-01-01 38 1 Ozone SAGUARO PA…
2 2017-01-02 40 2 Ozone SAGUARO PA…
3 2017-01-03 38 3 Ozone SAGUARO PA…
4 2017-01-04 38 4 Ozone SAGUARO PA…
5 2017-01-05 32 5 Ozone SAGUARO PA…
# ℹ 2,387 more rows
# ℹ 8 more variables: site_id <chr>, source <chr>,
# x20_year_high_2000_2019 <dbl>, x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>, good_aqi <dbl>Plot trend since 2016
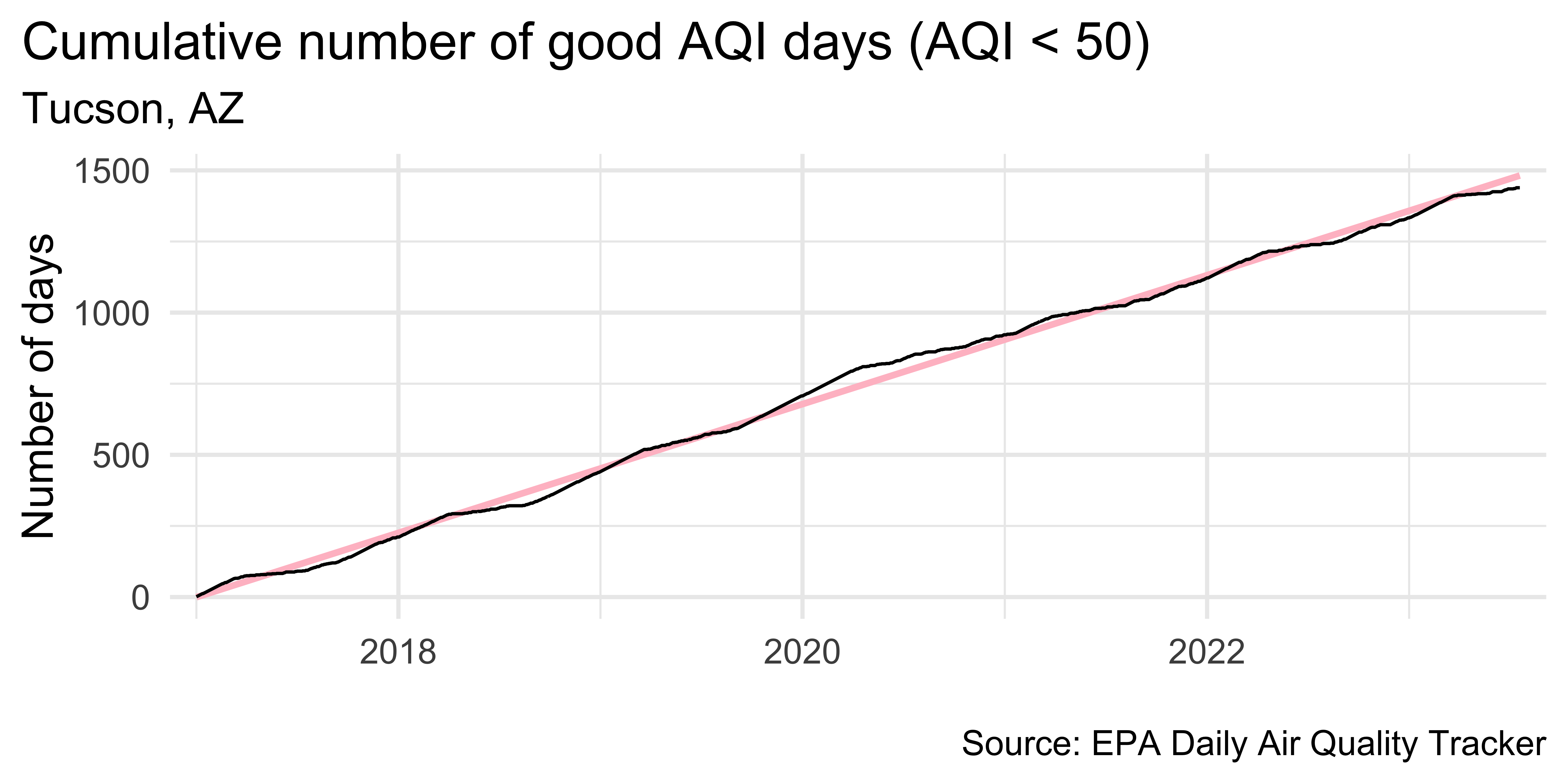
tuc |> ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) + geom_smooth(method = "lm", color = "pink") + geom_line() + scale_x_date( expand = expansion(mult = 0.02), date_labels = "%Y" ) + labs( x = NULL, y = "Number of days", title = "Cumulative number of good AQI days (AQI < 50)", subtitle = "Tucson, AZ", caption = "\nSource: EPA Daily Air Quality Tracker" ) + theme(plot.title.position = "plot")tuc |> ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) + geom_smooth(method = "lm", color = "pink") + geom_line() + scale_x_date( expand = expansion(mult = 0.02), date_labels = "%Y" ) + labs( x = NULL, y = "Number of days", title = "Cumulative number of good AQI days (AQI < 50)", subtitle = "Tucson, AZ", caption = "\nSource: EPA Daily Air Quality Tracker" ) + theme(plot.title.position = "plot")
`geom_smooth()` using formula = 'y ~ x'Detrend
Step 1. Fit a simple linear regression
Detrend
Step 2. Augment the data with model results (using broom::augment())
# A tibble: 2,392 × 8
cumsum_good_aqi date .fitted .resid .hat .sigma
<dbl> <date> <dbl> <dbl> <dbl> <dbl>
1 1 2017-01-01 -0.574 1.57 0.00167 23.9
2 2 2017-01-02 0.0457 1.95 0.00167 23.9
3 3 2017-01-03 0.666 2.33 0.00167 23.9
4 4 2017-01-04 1.29 2.71 0.00166 23.9
5 5 2017-01-05 1.91 3.09 0.00166 23.9
# ℹ 2,387 more rows
# ℹ 2 more variables: .cooksd <dbl>, .std.resid <dbl>Detrend
Step 3. Divide the observed value of cumsum_good_aqi by the respective value in the long-term trend (i.e., .fitted)
# A tibble: 2,392 × 9
cumsum_good_aqi date .fitted ratio .resid .hat .sigma
<dbl> <date> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 2017-01-01 -0.574 -1.74 1.57 0.00167 23.9
2 2 2017-01-02 0.0457 43.7 1.95 0.00167 23.9
3 3 2017-01-03 0.666 4.51 2.33 0.00167 23.9
4 4 2017-01-04 1.29 3.11 2.71 0.00166 23.9
5 5 2017-01-05 1.91 2.62 3.09 0.00166 23.9
# ℹ 2,387 more rows
# ℹ 2 more variables: .cooksd <dbl>, .std.resid <dbl>Visualize detrended data
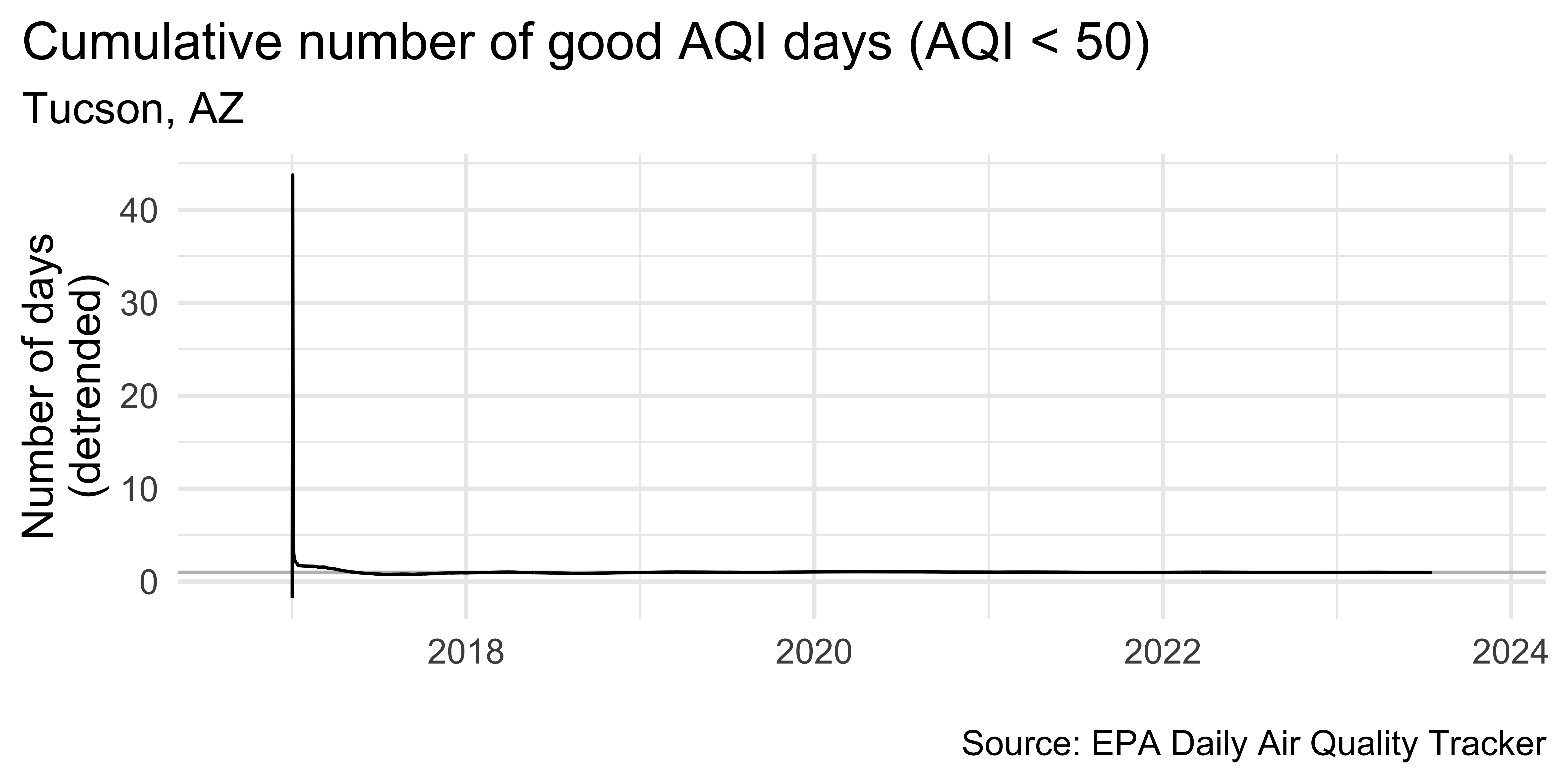
tuc_aug |>
ggplot(aes(x = date, y = ratio, group = 1)) +
geom_hline(yintercept = 1, color = "gray") +
geom_line() +
scale_x_date(
expand = expansion(mult = 0.1),
date_labels = "%Y"
) +
labs(
x = NULL, y = "Number of days\n(detrended)",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "Tucson, AZ",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")Air Quality in Tucson
barely anything interesting happening!
let’s look at data from somewhere with a bit more “interesting” air quality data…
Read in multiple years of SF data
sf <- read_csv(sf_files, na = c(".", ""))
sf <- sf |>
janitor::clean_names() |>
mutate(
date = mdy(date),
good_aqi = if_else(aqi_value <= 50, 1, 0)
) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(cumsum_good_aqi = cumsum(good_aqi), .after = aqi_value)
sf# A tibble: 2,557 × 13
date aqi_value cumsum_good_aqi main_pollutant site_name
<date> <dbl> <dbl> <chr> <chr>
1 2016-01-01 32 1 PM2.5 Durham Arm…
2 2016-01-02 37 2 PM2.5 Durham Arm…
3 2016-01-03 45 3 PM2.5 Durham Arm…
4 2016-01-04 33 4 PM2.5 Durham Arm…
5 2016-01-05 27 5 PM2.5 Durham Arm…
# ℹ 2,552 more rows
# ℹ 8 more variables: site_id <chr>, source <chr>,
# x20_year_high_2000_2019 <dbl>, x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>, good_aqi <dbl>Plot trend since 2016
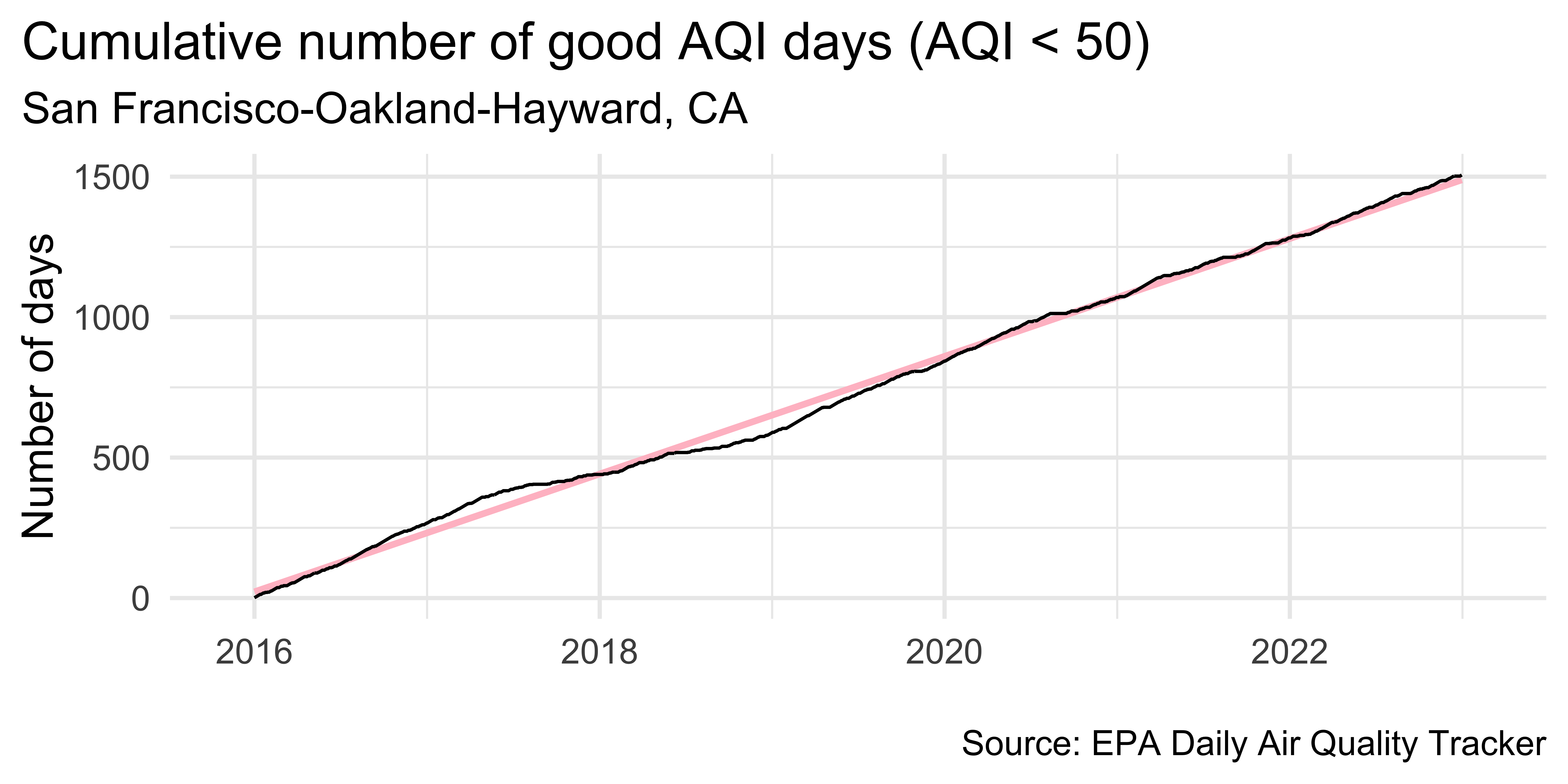
sf |>
ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) +
geom_smooth(method = "lm", color = "pink") +
geom_line() +
scale_x_date(
expand = expansion(mult = 0.07),
date_labels = "%Y"
) +
labs(
x = NULL, y = "Number of days",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "San Francisco-Oakland-Hayward, CA",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")`geom_smooth()` using formula = 'y ~ x'Detrend
- Fit a simple linear regression
Visualize detrended data
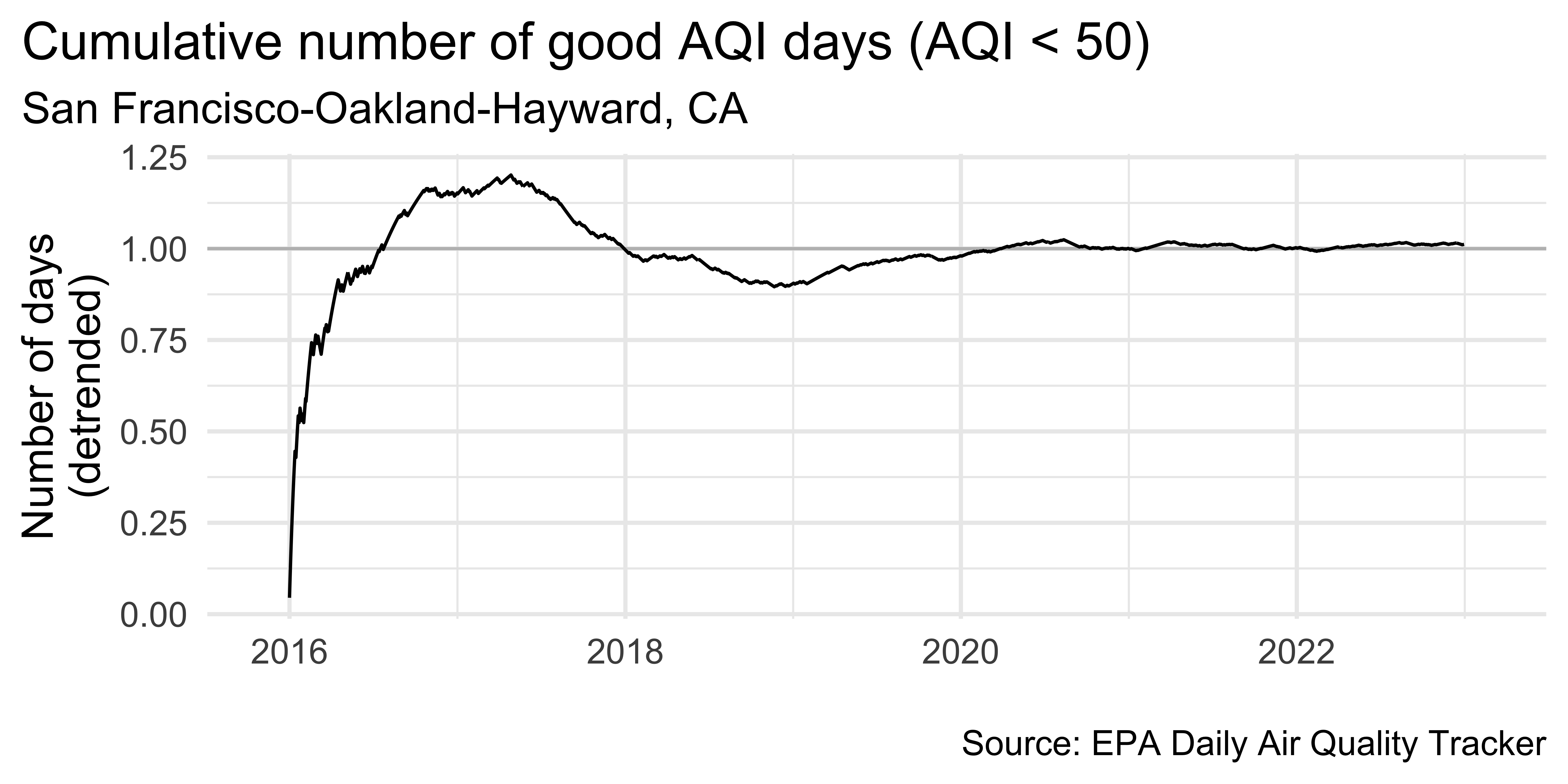
sf_aug |>
ggplot(aes(x = date, y = ratio, group = 1)) +
geom_hline(yintercept = 1, color = "gray") +
geom_line() +
scale_x_date(
expand = expansion(mult = 0.07),
date_labels = "%Y"
) +
labs(
x = NULL, y = "Number of days\n(detrended)",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "San Francisco-Oakland-Hayward, CA",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")Detrending
In step 2 we fit a very simple model
Depending on the complexity you’re trying to capture you might choose to fit a much more complex model
You can also decompose the trend into multiple trends, e.g. monthly, long-term, seasonal, etc.